SUIT-001 O2 mt D001: Difference between revisions
From Bioblast
No edit summary |
No edit summary |
||
| Line 22: | Line 22: | ||
:::: [[1D;2OctM;3P;4G;5S;6Gp;7U;8Rot-|SUIT RP2]]:1D;1M.1;2Oct;2c;2M2;3P;4G;5S;6Gp;7U;'''8Rot''';9Ama;'''10Tm''';11Azd | :::: [[1D;2OctM;3P;4G;5S;6Gp;7U;8Rot-|SUIT RP2]]:1D;1M.1;2Oct;2c;2M2;3P;4G;5S;6Gp;7U;'''8Rot''';9Ama;'''10Tm''';11Azd | ||
[[File:SUIT-Reference-Protocol.jpg|400px]] '''Harmonization between RP1 and RP2''' | |||
== DL-drivers for SUIT protocol RP1 == | == DL-drivers for SUIT protocol RP1 == | ||
Revision as of 10:26, 16 March 2017
Description
Abbreviation: FNSGp(PGM)
Reference: A: SUIT_RP1 - MiPNet21.06 SUIT RP
MitoPedia concepts:
SUIT protocol,
SUIT A
- MitoPedia: SUIT - SUIT reference protocol RP1
- SUIT-category: FNSGp(PGM)
- SUIT protocol pattern: orthogonal
Harmonized SUIT protocols
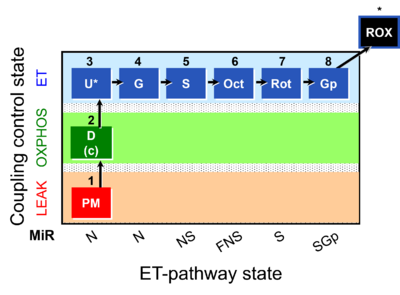
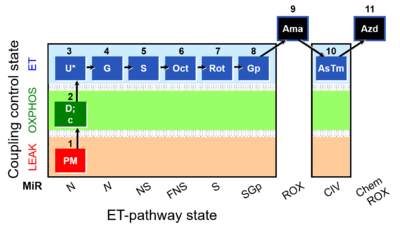
- SUIT RP1:1PM;2D:2c;3U;4G;5S;6Oct;7Rot;8Gp;9Ama;10Tm;11Azd
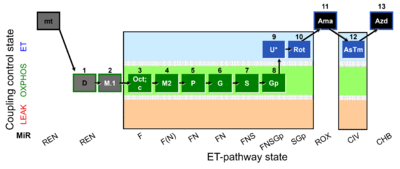
- SUIT RP2:1D;1M.1;2Oct;2c;2M2;3P;4G;5S;6Gp;7U;8Rot;9Ama;10Tm;11Azd
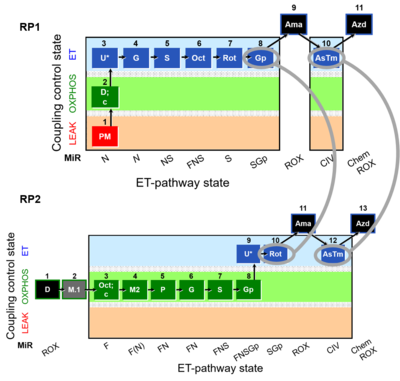 Harmonization between RP1 and RP2
Harmonization between RP1 and RP2
DL-drivers for SUIT protocol RP1
| SUIT Reference protocol | Sample | O2k-Core | O2k-Amp (H2O2 production) | ||
|---|---|---|---|---|---|
| Lab sheets | DL-Drivers version 7.1 | Lab sheets | DL-Drivers version 7.1 | ||
| RP1 | mt-preparations | »SUIT RP1mt | »RP1mt.XML | »SUIT RP1mt_AmR | In progress |
| RP1 | Permeabilized cells (pce) with digitonin | »SUIT RP1pce(Dig) | In progress | »SUIT RP1pce(Dig)_AmR | In progress |
| RP1 | Permeabilized cells (pce) without digitonin | »SUIT RP1pce | In progress | »SUIT RP1pce_AmR | In progress |
| RP1 | Permeabilized fibers | »SUIT RP1pfi | In progress | - | In progress |
DatLab-Analysis templates for RP1 analysis
| SUIT Reference protocol | Sample | O2k-Core | O2k-Amp (H2O2 production) |
|---|---|---|---|
| RP1 | mt-preparations | »SUIT RP1mt | - |
| RP1 | Permeabilized cells (pce) with digitonin | »SUIT RP1pce(Dig) | - |
| RP1 | Permeabilized cells (pce) without digitonin | »SUIT RP1pce | - |
| RP1 | Permeabilized fibers | »SUIT RP1mt | - |