Difference between revisions of "SUIT-001 O2 ce-pce D003"
From Bioblast
m (Iglesias-Gonzalez Javier moved page SUIT-001 O2 pce D03 to SUIT-001 O2 pce D003: New Nomenclature) |
|||
| Line 1: | Line 1: | ||
{{MitoPedia | {{MitoPedia | ||
|abbr=FNSGp(Oct,PGM) | |abbr=FNSGp(Oct,PGM) | ||
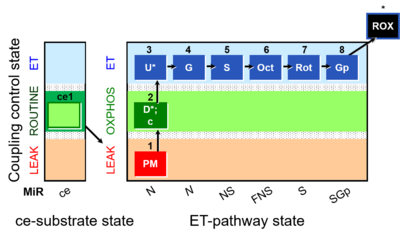
|description=[[File:1PM;2D;2c;3U;4G;5S;6Oct;7Rot;8Gp-.png|400px|SUIT-RP1]] | |description=[[File:ce1;1Dig;1PM;2D;2c;3U;4G;5S;6Oct;7Rot;8Gp-.png|400px|SUIT-RP1]] | ||
|info='''A: RP1''' - [[SUIT-001]] | |info='''A: RP1''' - [[SUIT-001]] | ||
|application=O2 | |application=O2 | ||
| Line 7: | Line 7: | ||
}} | }} | ||
::: [[MitoPedia: SUIT]] - '''[[SUIT reference protocol]] RP1''' | ::: [[MitoPedia: SUIT]] - '''[[SUIT reference protocol]] RP1''' | ||
::: '''[[Categories of SUIT protocols|SUIT-category]]:''' FNSGp(PGM) | ::: '''[[Categories of SUIT protocols |SUIT-category]]:''' FNSGp(Oct,PGM) | ||
::: '''[[SUIT protocol pattern]]:''' orthogonal 1PM;2D;2c;3U;4G;5S;6Oct;7Rot;8Gp | ::: '''[[SUIT protocol pattern]]:''' orthogonal 1PM;2D;2c;3U;4G;5S;6Oct;7Rot;8Gp;9Ama | ||
__TOC__ | __TOC__ | ||
== References == | == References == | ||
{{#ask:[[Category:Publications]] [[Additional label::SUIT- | {{#ask:[[Category:Publications]] [[Additional label::SUIT-001 O2 pce D003]] | ||
| ?Was published in year=Year | | ?Was published in year=Year | ||
| ?Has title=Reference | | ?Has title=Reference | ||
| Line 57: | Line 23: | ||
| order=descending | | order=descending | ||
}} | }} | ||
{{Template:SUIT-001}} | |||
== Strengths and limitations == | |||
:::* SUIT-001 in combination with [[SUIT-002]] provides a common reference for comparison of respiratory control in a large variety of species, tissues and cell types. Both SUIT protocols provide a mitochondrial mapping which allows: | |||
:::: 1. to obtain reference values. | |||
:::: 2. to evaluate mitochondrial physiological diversity, generating a mt-database on comparative mitochondrial physiology. | |||
:::: 3. to screen specific defects. | |||
:::* SUIT-001 and [[SUIT-002]] are used in the [[Proficiency_test| MitoFit Proficiency test]] for inter-individual and inter-laboratory reproducibility quality control. | |||
:::* A succinate concentration of >10 mM may be required for saturating S<sub>''E''</sub> capacity. | |||
:::+ Linear coupling control (''L''-''P''-''E'') with N substrates (PM). | |||
:::+ Pathway control in ET state (N, NS, FNS, S and SGp pathways). | |||
:::+ Harmonization with [[SUIT-002]] allows to perform both SUIT protocols in parallel. The cross-linked respiratory states can be statically used as repeat measurements. | |||
:::+ In cells, SUIT-001 and SUIT-002 harmonize in: [[ROUTINE]] (ce1), SGp<sub>''E''</sub> (8Gp in SUIT-001 and 10Rot in SUIT-002) and Complex IV. | |||
:::+ Harmonization with many SUIT protocols (up to step 5S). | |||
:::+ Oct is added in ET state to avoid uncoupler effect with fatty acids. | |||
:::+ If NS- and FNS-pathways are similar (NS<sub>''E''</sub> ~ FNS<sub>''E''</sub>), the additivity index of N- and S-pathways in ET state can be evaluated. | |||
:::+ This protocol can be extended with the Complex IV module. | |||
:::- The substrate combination for maximum ET capacity (FNSGP) is not obtained in SUIT-001 in favour of measuring S<sub>''E''</sub>. | |||
== Compare SUIT protocols == | |||
::::* [[SUIT-002 O2 pce D007]] (RP2): Harmonized SUIT protocol for permeabilized cells. | |||
::::* [[SUIT-004]]: Short version of RP1; without G, F and Gp. | |||
::::* [[SUIT-006]]: Very short initial coupling control module. | |||
| Line 65: | Line 62: | ||
|mitopedia method=Respirometry | |mitopedia method=Respirometry | ||
}} | }} | ||
{{Labeling | |||
|additional=SUIT-001}} | |||
[[Category:Publications]] | |||
Revision as of 13:22, 21 January 2019
Description
Abbreviation: FNSGp(Oct,PGM)
Reference: A: RP1 - SUIT-001
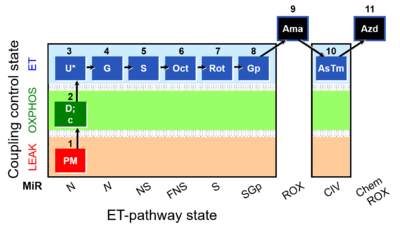
SUIT number: D003_ce1;1Dig;1PM;2D;2c;3U;4G;5S;6Oct;7Rot;8Gp;9Ama;10AsTm;11Azd
O2k-Application: O2
- MitoPedia: SUIT - SUIT reference protocol RP1
- SUIT-category: FNSGp(Oct,PGM)
- SUIT protocol pattern: orthogonal 1PM;2D;2c;3U;4G;5S;6Oct;7Rot;8Gp;9Ama
References
Steps and respiratory states
| Step | State | Pathway | Q-junction | Comment - Events (E) and Marks (M) |
|---|---|---|---|---|
| 1PM | PML(n) | N | CI | 1PM
|
| 2D | PMP | N | CI | 1PM;2D
|
| 2c | PMcP | N | CI | 1PM;2D;2c
|
| 3U | PME | N | CI | 1PM;2D;3U
|
| 4G | PGME | N | CI | 1PM;2D;3U;4G
|
| 5S | PGMSE | NS | CI&II | 1PM;2D;3U;4G;5S
|
| 6Oct | OctPGMSE | FNS | FAO&CI&II | 1PM;2D;3U;4G;5S;6Oct
|
| 7Rot | SE | S | CII | 1PM;2D;3U;4G;5S;6Oct;7Rot
|
| 8Gp | SGpE | SGp | CII&GpDH | 1PM;2D;3U;4G;5S;6Oct;7Rot;8Gp
|
| 9Ama | ROX | 1PM;2D;3U;4G;5S;6Oct;7Rot;8Gp;9Ama
|
| Step | Respiratory state | Pathway control | ET-Complex | Comment |
|---|---|---|---|---|
| ## AsTm | AsTmE | CIV | CIV | |
| ## Azd | CHB |
- Bioblast links: SUIT protocols - >>>>>>> - Click on [Expand] or [Collapse] - >>>>>>>
- Coupling control
- Pathway control
- Main fuel substrates
- » Glutamate, G
- » Glycerophosphate, Gp
- » Malate, M
- » Octanoylcarnitine, Oct
- » Pyruvate, P
- » Succinate, S
- Main fuel substrates
- Glossary
Strengths and limitations
- SUIT-001 in combination with SUIT-002 provides a common reference for comparison of respiratory control in a large variety of species, tissues and cell types. Both SUIT protocols provide a mitochondrial mapping which allows:
- 1. to obtain reference values.
- 2. to evaluate mitochondrial physiological diversity, generating a mt-database on comparative mitochondrial physiology.
- 3. to screen specific defects.
- SUIT-001 and SUIT-002 are used in the MitoFit Proficiency test for inter-individual and inter-laboratory reproducibility quality control.
- A succinate concentration of >10 mM may be required for saturating SE capacity.
- + Linear coupling control (L-P-E) with N substrates (PM).
- + Pathway control in ET state (N, NS, FNS, S and SGp pathways).
- + Harmonization with SUIT-002 allows to perform both SUIT protocols in parallel. The cross-linked respiratory states can be statically used as repeat measurements.
- + In cells, SUIT-001 and SUIT-002 harmonize in: ROUTINE (ce1), SGpE (8Gp in SUIT-001 and 10Rot in SUIT-002) and Complex IV.
- + Harmonization with many SUIT protocols (up to step 5S).
- + Oct is added in ET state to avoid uncoupler effect with fatty acids.
- + If NS- and FNS-pathways are similar (NSE ~ FNSE), the additivity index of N- and S-pathways in ET state can be evaluated.
- + This protocol can be extended with the Complex IV module.
- - The substrate combination for maximum ET capacity (FNSGP) is not obtained in SUIT-001 in favour of measuring SE.
Compare SUIT protocols
- SUIT-002 O2 pce D007 (RP2): Harmonized SUIT protocol for permeabilized cells.
MitoPedia concepts: SUIT protocol, SUIT A, Find
MitoPedia methods:
Respirometry
Labels:
SUIT-001