Difference between revisions of "PM-pathway control state"
From Bioblast
Beno Marija (talk | contribs) |
|||
| Line 15: | Line 15: | ||
[[File:PM.jpg|right|400px|link=Gnaiger 2014 MitoPathways |Gnaiger 2014 MitoPathways - Chapter 3.2]] | [[File:PM.jpg|right|400px|link=Gnaiger 2014 MitoPathways |Gnaiger 2014 MitoPathways - Chapter 3.2]] | ||
== PM<sub>''L''</sub> == | == PM<sub>''L''</sub> == | ||
::::* [[ | ::::* [[SUIT-1 O2 mt D01]] | ||
::::* [[SUIT-1 O2 pfi D02]] | |||
::::* [[SUIT-1 O2 pce D03]] | |||
::::* [[SUIT-1 O2 pce D04]] | |||
::::* [[SUIT-4 O2 pfi D10]] | |||
::::* [[SUIT-8 O2 pfi D14]] | |||
== PM<sub>''P''</sub> == | == PM<sub>''P''</sub> == | ||
::::* [[ | ::::* [[SUIT-1 O2 mt D01]] | ||
::::* [[SUIT-1 O2 pfi D02]] | |||
::::* [[SUIT-1 O2 pce D03]] | |||
::::* [[SUIT-1 O2 pce D04]] | |||
::::* [[SUIT-4 O2 pfi D10]] | |||
::::* [[SUIT-8 O2 pfi D14]] | |||
== PM<sub>''E''</sub> == | == PM<sub>''E''</sub> == | ||
::::* [[ | ::::* [[SUIT-1 O2 mt D01]] | ||
::::* [[SUIT-1 O2 pfi D02]] | |||
::::* [[SUIT-1 O2 pce D03]] | |||
::::* [[SUIT-1 O2 pce D04]] | |||
::::* [[SUIT-4 O2 pfi D10]] | |||
Revision as of 17:23, 9 January 2019
Description
MitoPathway control state: N
SUIT protocol: 1PM;2D;3U;4G;5S;6Oct;7Rot;8Gp- - SUIT_RP1
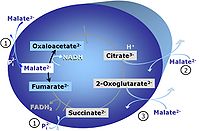
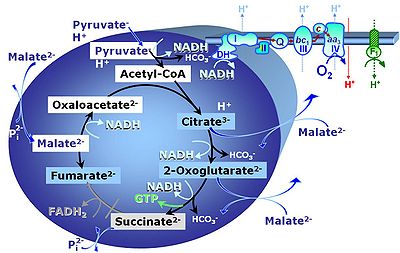
Pyruvate (P) is oxidatively decarboxylated to acetyl-CoA and CO2, yielding NADH catalyzed by pyruvate dehydrogenase. Malate (M) is oxidized to oxaloacetate by mt-malate dehydrogenase located in the mitochondrial matrix. Condensation of oxaloacate with acetyl-CoA yields citrate (citrate synthase). 2-oxoglutarate (α-ketoglutarate) is formed from isocitrate (isocitrate dehydrogenase).
Abbreviation: PM
Reference: Gnaiger 2014 MitoPathways - Chapter 3.2
MitoPedia concepts:
SUIT state
PML
PMP
PME
Linear coupling control in the N-pathway control state: L – P - E
- L - P
- OXPHOS coupling efficiency (P-L or ≈P control factor), j≈P = ≈P/P = (P-L)/P = 1-L/P, is measured in the CI-linked substrate state, with defined coupling sites (CI, CIII, CIV) and at high flux.
- P - E
- CCCP is titrated stepwise to maximum flux, to evaluate limitation of OXPHOS by the phosphorylation system, expressed as the apparent excess E-P capacity factor (E-P coupling control factor), jExP = (E-P)/E = 1-P/E. If jExP>0, then the ET-coupling efficiency rather than the OXPHOS coupling efficiency is the proper expression of coupling, j≈E = ≈E/E = (E-L)/E = 1-L/E.
Discussion
- The Pyruvate anaplerotic pathway control state (pyruvate alone) is not an ET-pathway competent substrate state in most mt-preparations, since acetyl-CoA accumulates without the co-substrate (oxaloacetate) of citrate synthase.
- The Malate anaplerotic pathway control state (M alone) is not an ET-pathway competent substrate state in many mt-preparations, since oxaloacetate accumulates without the co-substrate (acetyl-CoA) of citrate synthase.