Difference between revisions of "P-L net OXPHOS capacity"
| Line 1: | Line 1: | ||
{{MitoPedia | {{MitoPedia | ||
|abbr=''≈P'' | |abbr=''≈P'' | ||
|description=[[File:EPL-free and excess.jpg|left| | |description=[[File:EPL-free and excess.jpg|left|240px|thumb|Blue Book 2014: Fig. 2.4.]] | ||
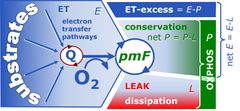
The '''free OXPHOS capacity''', ''≈P'', is the [[OXPHOS capacity]] corrected for [[LEAK respiration]], ''≈P'' = ''P-L''. ''≈P'' is the respiratory capacity potentially available for phosphorylation of ADP to ATP. Oxygen consumption in the OXPHOS state, therefore, is partitioned into the free OXPHOS capacity, ''≈P'', strictly coupled to phosphorylation, ''~P'', and nonphosphorylating LEAK respiration, ''L<sub>P</sub>'', compensating for proton leaks, slip and cation cycling: ''P'' = ''≈P''+''L<sub>P</sub>''. It is frequently assumed that [[LEAK respiration]], ''L'', as measured in the LEAK state, overestimates the LEAK component of respiration, ''L<sub>P</sub>'', as measured in the OXPHOS state, particularly if the protonmotive force is not adjusted to equivalent levels in ''L'' and ''L<sub>P</sub>''. However, if the LEAK component increases with enzyme turnover during ''P'', the low enzyme turnover during ''L'' may counteract the effect of the higher Δ''p''<sub>mt</sub>. | The '''free OXPHOS capacity''', ''≈P'', is the [[OXPHOS capacity]] corrected for [[LEAK respiration]], ''≈P'' = ''P-L''. ''≈P'' is the respiratory capacity potentially available for phosphorylation of ADP to ATP. Oxygen consumption in the OXPHOS state, therefore, is partitioned into the free OXPHOS capacity, ''≈P'', strictly coupled to phosphorylation, ''~P'', and nonphosphorylating LEAK respiration, ''L<sub>P</sub>'', compensating for proton leaks, slip and cation cycling: ''P'' = ''≈P''+''L<sub>P</sub>''. It is frequently assumed that [[LEAK respiration]], ''L'', as measured in the LEAK state, overestimates the LEAK component of respiration, ''L<sub>P</sub>'', as measured in the OXPHOS state, particularly if the protonmotive force is not adjusted to equivalent levels in ''L'' and ''L<sub>P</sub>''. However, if the LEAK component increases with enzyme turnover during ''P'', the low enzyme turnover during ''L'' may counteract the effect of the higher Δ''p''<sub>mt</sub>. | ||
|info=[[Gnaiger 2014 MitoPathways]] | |info=[[Gnaiger 2014 MitoPathways]] | ||
Revision as of 16:21, 21 September 2014
Description
The free OXPHOS capacity, ≈P, is the OXPHOS capacity corrected for LEAK respiration, ≈P = P-L. ≈P is the respiratory capacity potentially available for phosphorylation of ADP to ATP. Oxygen consumption in the OXPHOS state, therefore, is partitioned into the free OXPHOS capacity, ≈P, strictly coupled to phosphorylation, ~P, and nonphosphorylating LEAK respiration, LP, compensating for proton leaks, slip and cation cycling: P = ≈P+LP. It is frequently assumed that LEAK respiration, L, as measured in the LEAK state, overestimates the LEAK component of respiration, LP, as measured in the OXPHOS state, particularly if the protonmotive force is not adjusted to equivalent levels in L and LP. However, if the LEAK component increases with enzyme turnover during P, the low enzyme turnover during L may counteract the effect of the higher Δpmt.
Abbreviation: ≈P
Reference: Gnaiger 2014 MitoPathways
MitoPedia methods:
Respirometry
MitoPedia topics: "Respiratory state" is not in the list (Enzyme, Medium, Inhibitor, Substrate and metabolite, Uncoupler, Sample preparation, Permeabilization agent, EAGLE, MitoGlobal Organizations, MitoGlobal Centres, ...) of allowed values for the "MitoPedia topic" property.
Respiratory state"Respiratory state" is not in the list (Enzyme, Medium, Inhibitor, Substrate and metabolite, Uncoupler, Sample preparation, Permeabilization agent, EAGLE, MitoGlobal Organizations, MitoGlobal Centres, ...) of allowed values for the "MitoPedia topic" property.
Compare
- Free ROUTINE activity (intact cells)
- Free ETS capacity