Difference between revisions of "MiPNet21.06 SUIT RP"
| Line 245: | Line 245: | ||
| | | | ||
| pce | | pce | ||
| | | | ||
| | | | ||
| | | | ||
|- | |- | ||
| Titration sheet | | Titration sheet | ||
Revision as of 22:59, 19 November 2016
| SUIT reference protocol for OXPHOS analysis by high-resolution respirometry. |
OROBOROS (2016-08-17) Mitochondr Physiol Network
Abstract: Doerrier C, Sumbalova Z, Krumschnabel G, Hiller E, Gnaiger (2016) SUIT reference protocol for OXPHOS analysis by high-resolution respirometry. Mitochondr Physiol Network 21.06(01):1-12. » Versions
- » MitoPedia: SUIT reference protocol
- » Instrument: O2k, O2k-Catalogue
• O2k-Network Lab: AT_Innsbruck_OROBOROS
Labels: MiParea: Instruments;methods
Preparation: Permeabilized cells, Permeabilized tissue, Homogenate, Isolated mitochondria
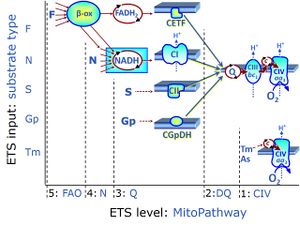
Regulation: Substrate Coupling state: LEAK, OXPHOS, ETS"ETS" is not in the list (LEAK, ROUTINE, OXPHOS, ET) of allowed values for the "Coupling states" property. Pathway: F, N, S, Gp, CIV, NS, Other combinations HRR: Oxygraph-2k, O2k-Protocol
MitoPathways, MitoFitPublication
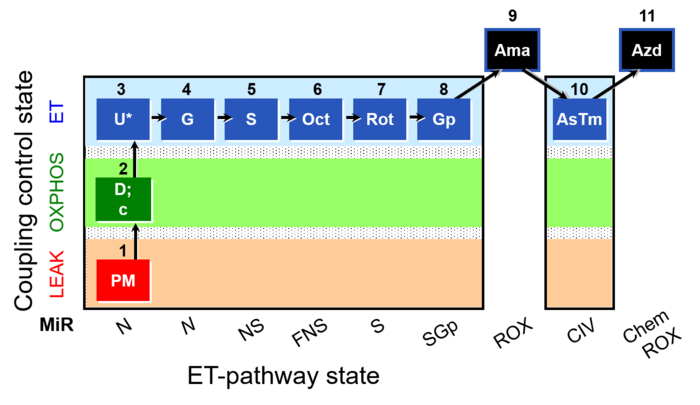
SUIT reference protocol-RP1
SUIT states: 1-4PM(LPcE) 5PGM 6PGMS 7OctPGMS 8S 9SGp 10ROX 11Tm 12ROX
| Step | Respiratory state | Pathway control | Pathway to Q |
|---|---|---|---|
| 1PM | PM(L) | N | CI |
| 2D | PM(P) | N | CI |
| 3c | PM(P)c | N | CI |
| 3NADH | PM(P)cNADH | N | CI |
| 4U | PM(E) | N | CI |
| 5G | PGM(E) | N | CI |
| 6S | PGMS(E) | NS | CI&II |
| 7Oct | OctPGMS(E) | FNS | FAO&CI&II |
| 8Rot | S(E) | S | CII |
| 9Gp | SGp(E) | SGp | CII&GpDH |
| 10Ama | ROX | ROX | |
| 11Tm | Tm(E) | CIV | CIV |
| 12Azd | ROX | ROX |
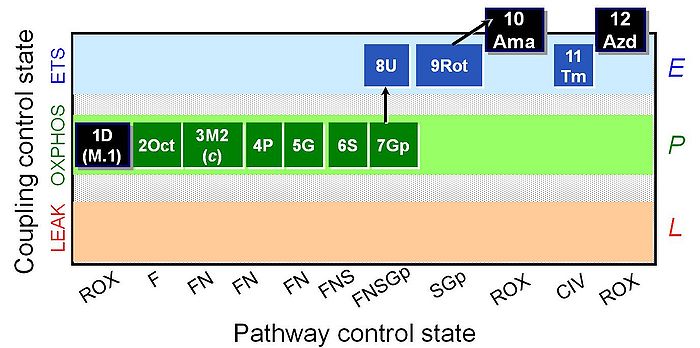
SUIT reference protocol-RP2
SUIT states: 1ROX 2Oct(P) 3OctM(4c) 5OctPM 6OctPGM 7OctPGMS 8OctPGMSGp 9E 10SGp 11ROX 12CIV 13ROX
| Step | Respiratory state | Pathway control | Pathway to Q |
|---|---|---|---|
| 1D | ROX | ROX | |
| 2Oct | Oct(P) | (F) | FAO |
| 3Mtit | OctM(P) | F | FAO |
| 4c | OctM(P)c | F | FAO |
| 4NADH | OctM(P)cNADH | F | FAO |
| 5P | OctPM(P) | FN | FAO&CI |
| 6G | OctPGM(P) | FN | FAO&CI |
| 7S | OctPGMS(P) | FNS | FAO&CI&II |
| 8Gp | OctPGMSGp(P) | FNSGp | FAO&CI&II&GpDH |
| 9U | OctPGMSGp(E) | FNSGp | FAO&CI&II&GpDH |
| 10Rot | SGp(E) | SGp | CII&GpDH |
| 11Ama | ROX | ROX | |
| 12Tm | Tm(E) | CIV | CIV |
| 13Azd | ROX | ROX |
Laboratory sheets for SUIT protocols RP1 and RP2
| Type | SUIT protocol | Sample | Abbr | docx file | pdf file | Last update |
|---|---|---|---|---|---|---|
| Titration sheet | RP1 | Permeabilized cells | pce | »SUIT_RP1pce | »SUIT_RP1pce | 2016-11-19 |
| Titration sheet | RP2 | pce | »SUIT_RP2pce | »SUIT_RP2pce | 2016-11-19 | |
| Tick boxes | Combined | pce | ||||
| Titration sheet | RP1_AmR | pce | »SUIT_RP1_AmR_pce | »SUIT_RP1_AmR_pce | 2016-11-19 | |
| Titration sheet | RP2_AmR | pce | »SUIT_RP2_AmR_pce | »SUIT_RP2_AmR_pce | 2016-11-19 | |
| Titration sheet | RP1 | Isolated mitochondria, tissue homogenate, permeabilized fibers | imt, thom, pfi | |||
| Titration sheet | RP2 | imt, thom, pfi | ||||
| Tick boxes | Combined | imt, thom, pfi |
DatLab-Analysis templates for RP analysis
Experimental details
Work in progress: pre-publication protocol
Saturating ADP concentrations
- Different concentrations of ADP are sufficient to obtain maximum flux for estimating OXPHOS capacity. The typical range for isolated mitochondria is 1 to 2.5 mM, for permeabilized cells 1 to 5 mM, for permeabilized muscle fibres 2.5 to 10 mM.
RP-T01
- RP1: Compared to RP1-T01, move Oct titration after S.
- RP2: Compared to RP2-T01, move U titration after Gp.
Cytochrome c test
- In protocol RP2, the cytochrome c test might better be moved from a c-titration after pyruvate (P) to a titration after M2. Otherwise, there is a risk of underestimation of FAO in cases of a cytochrome c effect. On the other hand, at the low flux before titration of P, the sensitivity of the cytochrome c test may be low.
- ~ Gnaiger Erich 18:26, 23 January 2016 (CET)
Mark names
- Even the highly abbreviated names of respiratory states have become too long for DatLab, where mark names are restricted to 8 digits. The visibility in DatLab of long mark names is restricted. The simplest solution for short mark names is the use of a numerical sequence, with the preceeding event name added for information.
- ~ Gnaiger Erich 16:50, 23 January 2016 (CET)
Pre-publication communication
- This communication is a pre-publication, inviting critical feedback and comments. All feedback will be carefully documented and evaluated in terms of justification of co-authorship. We intend to finally publish this topic in a peer-reviewed journal as an original article, with reference to the pre-publication history including reviewer's reports. - Gnaiger Erich 03:23, 18 January 2016 (CET)
Experiments in progress
- The SUIT reference protocol is presently applied in permeabilized HEK cells, mouse heart isolated mitochondria, liver homogenate, permeabilized skeletal muscle (mouse and human), and human PBMCs and platelets. - Gnaiger Erich 18:47, 19 January 2016 (CET)
- AT Innsbruck Gnaiger E and AT Innsbruck OROBOROS - MitoFit project.
- US NC Winston-Salem Molina AJA - UPBEAT project.
Further details
- » Introduction: Gnaiger 2014 MitoPathways
- » MitoPedia: SUIT - extended MitoPedia topic, May 2016
- Table of titrations » MiPNet09.12 O2k-Titrations
- » Definition: Substrate-uncoupler-inhibitor titration
- » Context: SUIT protocol library
- » Abbreviations: MitoPedia