MiPNet21.06 SUIT RP: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
{{Publication | {{Publication | ||
|title=[[Image:O2k-Protocols.jpg|right|80px|link=O2k-Protocols|O2k-Protocols]] SUIT reference | |title=[[Image:O2k-Protocols.jpg|right|80px|link=O2k-Protocols|O2k-Protocols]] SUIT reference protocol for OXPHOS analysis by high-resolution respirometry. - [[Media:MiPNet21.06 SUIT reference protocol.pdf |»Bioblast pdf«]] | ||
|info=[http://www.bioblast.at/index.php/File:MiPNet21. | |info=[http://www.bioblast.at/index.php/File:MiPNet21.06_SUIT_reference_protocol.pdf Versions] | ||
|authors=OROBOROS | |authors=OROBOROS | ||
|year=2016-01-23 | |year=2016-01-23 | ||
|journal=Mitochondr Physiol Network | |journal=Mitochondr Physiol Network | ||
|abstract='''Doerrier C, Sumbalova Z, Lamberti G, Krumschnabel G, Gnaiger (2016) SUIT reference | |abstract='''Doerrier C, Sumbalova Z, Lamberti G, Krumschnabel G, Gnaiger (2016) SUIT reference protocol for OXPHOS analysis by high-resolution respirometry. Mitochondr Physiol Network 21.06(pre03):1-10.''' | ||
:» MitoPedia: [[SUIT reference | :» MitoPedia: [[SUIT reference protocol]] | ||
:» Instrument: [[Oxygraph-2k|O2k]], [[OROBOROS O2k-Catalogue | O2k-Catalogue]] | :» Instrument: [[Oxygraph-2k|O2k]], [[OROBOROS O2k-Catalogue | O2k-Catalogue]] | ||
|mipnetlab=AT_Innsbruck_OROBOROS | |mipnetlab=AT_Innsbruck_OROBOROS | ||
| Line 22: | Line 22: | ||
== Work in progress == | == Work in progress == | ||
== RP == | |||
* RP1: Compared to RP1-T01, move Oct titration after S. | |||
* RP2: Compared to RP2-T01, move U titration after Gp. | |||
== RP-T01 == | |||
=== Cytochrome c test === | === Cytochrome c test === | ||
* In protocol RP2, the cytochrome ''c'' test might better be moved from a c-titration after pyruvate (P) to a titration after M2. Otherwise, there is a risk of underestimation of FAO in cases of a cytochrome c effect. On the other hand, at the low flux before titration of P, the sensitivity of the cytochrome ''c'' test may be low. | * In protocol RP2, the cytochrome ''c'' test might better be moved from a c-titration after pyruvate (P) to a titration after M2. Otherwise, there is a risk of underestimation of FAO in cases of a cytochrome c effect. On the other hand, at the low flux before titration of P, the sensitivity of the cytochrome ''c'' test may be low. | ||
| Line 123: | Line 129: | ||
== Experimental details == | == Experimental details == | ||
''Work in progress towards the next pre-publication version of'' [[Media:MiPNet21.06 SUIT reference | ''Work in progress towards the next pre-publication version of'' [[Media:MiPNet21.06 SUIT reference protocol.pdf |»MiPNet21.06 SUIT reference protocol.pdf«]] | ||
=== Saturating ADP concentrations === | === Saturating ADP concentrations === | ||
| Line 133: | Line 139: | ||
=== Experiments in progress === | === Experiments in progress === | ||
The '''SUIT reference | The '''SUIT reference protocol''' is presently applied in permeabilized HEK cells, mouse heart isolated mitochondria, liver homogenate, permeabilized skeletal muscle (mouse and human), and human PBMCs and platelets. - [[User:Gnaiger Erich|Gnaiger Erich]] 18:47, 19 January 2016 (CET) | ||
* [[AT Innsbruck Gnaiger E]] and [[AT Innsbruck OROBOROS]] - [[O2k-MitoFit |MitoFit]] project. | * [[AT Innsbruck Gnaiger E]] and [[AT Innsbruck OROBOROS]] - [[O2k-MitoFit |MitoFit]] project. | ||
* [[US NC Winston-Salem Molina AJA]] - UPBEAT project. | * [[US NC Winston-Salem Molina AJA]] - UPBEAT project. | ||
Revision as of 23:15, 9 February 2016
| SUIT reference protocol for OXPHOS analysis by high-resolution respirometry. - »Bioblast pdf« |
» Versions
OROBOROS (2016-01-23) Mitochondr Physiol Network
Abstract: Doerrier C, Sumbalova Z, Lamberti G, Krumschnabel G, Gnaiger (2016) SUIT reference protocol for OXPHOS analysis by high-resolution respirometry. Mitochondr Physiol Network 21.06(pre03):1-10.
- » MitoPedia: SUIT reference protocol
- » Instrument: O2k, O2k-Catalogue
• O2k-Network Lab: AT_Innsbruck_OROBOROS
Labels: MiParea: Instruments;methods
Preparation: Permeabilized cells, Permeabilized tissue, Homogenate, Isolated mitochondria
Regulation: Substrate Coupling state: LEAK, OXPHOS, ETS"ETS" is not in the list (LEAK, ROUTINE, OXPHOS, ET) of allowed values for the "Coupling states" property.
HRR: Oxygraph-2k, O2k-Protocol
MitoPathways, MitoFitPublication
Work in progress
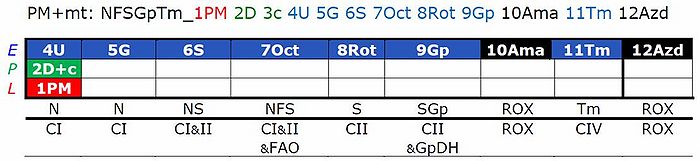
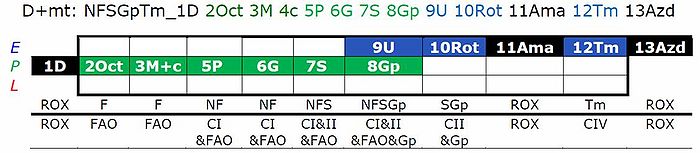
RP
- RP1: Compared to RP1-T01, move Oct titration after S.
- RP2: Compared to RP2-T01, move U titration after Gp.
RP-T01
Cytochrome c test
- In protocol RP2, the cytochrome c test might better be moved from a c-titration after pyruvate (P) to a titration after M2. Otherwise, there is a risk of underestimation of FAO in cases of a cytochrome c effect. On the other hand, at the low flux before titration of P, the sensitivity of the cytochrome c test may be low.
- ~ Gnaiger Erich 18:26, 23 January 2016 (CET)
Mark names
- Even the highly abbreviated names of respiratory states have become too long for DatLab, where mark names are restricted to 8 digits. The visibility in DatLab of long mark names is restricted. The simplest solution for short mark names is the use of a numerical sequence, with the preceeding event name added for information.
- ~ Gnaiger Erich 16:50, 23 January 2016 (CET)
Laboratory sheets for SUIT protocols RP1 and RP2
| Type | SUIT protocol | Sample | Abbr | docx file | pdf file | Last update |
|---|---|---|---|---|---|---|
| Titration sheet | RP1 | Permeabilized fibres | Pfi | »SUIT_RP1-Pfi | »SUIT_RP1-Pfi | 2016-01-30 |
| Titration sheet | RP2 | Pfi | »SUIT_RP2-Pfi | »SUIT_RP2-Pfi | 2016-01-30 | |
| Tick boxes | Combined | Pfi | »RP print-Pfi | »RP print-Pfi | 2016-01-30 | |
| Titration sheet | RP1 | Permeabilized cells | Pc | »SUIT_RP1-Pc | »SUIT_RP1-Pc | 2016-01-25 |
| Titration sheet | RP2 | Pc | »SUIT_RP2-Pc | »SUIT_RP2-Pc | 2016-01-25 | |
| Tick boxes | Combined | Pc | »RP-Pc print | »RP-Pc print | ||
| Titration sheet | RP1 | Isolated mitochondria, tissue homogenate | Imt, Thom | »SUIT_RP1-Imt | »SUIT_RP1-Imt | 2016-01-25 |
| Titration sheet | RP2 | Imt, Thom | »SUIT_RP2-Imt | »SUIT_RP2-Imt | 2016-01-25 | |
| Tick boxes | Combined | Imt, Thom | »RP print | »RP print |
Experimental details
Work in progress towards the next pre-publication version of »MiPNet21.06 SUIT reference protocol.pdf«
Saturating ADP concentrations
Different concentrations of ADP are sufficient to obtain maximum flux for estimating OXPHOS capacity. The typical range for isolated mitochondria is 1 to 2.5 mM, for permeabilized cells 1 to 5 mM, for permeabilized muscle fibres 2.5 to 10 mM.
Pre-publication communication
This communication is a pre-publication, inviting critical feedback and comments. All feedback will be carefully documented and evaluated in terms of justification of co-authorship. We intend to finally publish this topic in a peer-reviewed journal as an original article, with reference to the pre-publication history including reviewer's reports. - Gnaiger Erich 03:23, 18 January 2016 (CET)
Experiments in progress
The SUIT reference protocol is presently applied in permeabilized HEK cells, mouse heart isolated mitochondria, liver homogenate, permeabilized skeletal muscle (mouse and human), and human PBMCs and platelets. - Gnaiger Erich 18:47, 19 January 2016 (CET)
- AT Innsbruck Gnaiger E and AT Innsbruck OROBOROS - MitoFit project.
- US NC Winston-Salem Molina AJA - UPBEAT project.
Further details
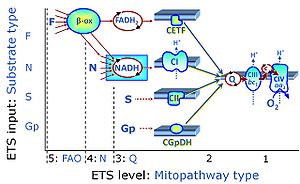
- Introduction » Gnaiger 2014 MitoPathways
- » MitoPedia: SUIT protocols - extended MitoPedia topic, Jan 2016
- » MitoPedia: SUIT states - extended MitoPedia topic, Jan 2016
- Table of titrations » MiPNet09.12 O2k-Titrations
- Definition » Substrate-uncoupler-inhibitor titration
- Context » SUIT protocol library
- Abbreviations » MitoPedia