Oliveira 2015 Abstract IOC100
| Oliveira MT, Saari S, Andjelkovic A, Jacobs HT (2015) Investigating the metabolic alterations caused by the transgenic expression of the mitochondrial alternative oxidase of Ciona intestinalis in Drosophila melanogaster. Mitochondr Physiol Network 20.01. |
Link:
Oliveira MT, Saari S, Andjelkovic A, Jacobs HT (2015)
Event: IOC100
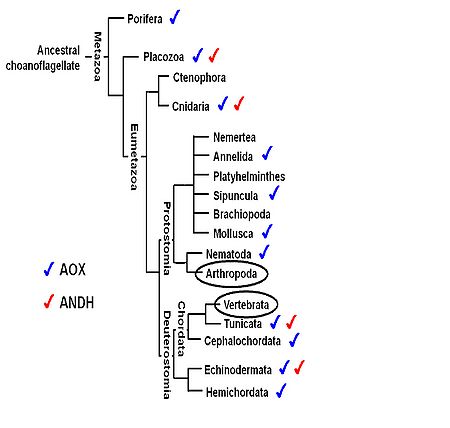
The respiratory chain alternative enzymes constitute additional pathways to the mitochondrial oxidative phosphorylation (OXPHOS) system, in which oxygen consumption is uncoupled from ATP production. The expression of alternative oxidases and alternative NADH dehydrogenases from tunicates and fungi in the fruitfly Drosophila melanogaster, in the mouse and in cultured human cells has proven to be benign and to counteract deleterious effects of defective OXPHOS systems, such as the high production of reactive oxygen species. The observed benefits are in paradox with the fact that genes for alternative enzymes were lost independently, early in the evolution of Vertebrata and Arthropoda (Figure 1). We thus propose to investigate the functions of an alternative enzyme from Ciona intestinalis (Tunicata: Ascidiacea), the alternative oxidase (AOX), when expressed in transgenic fruitflies. In preliminary thermal and nutritional stress experiments that challenge mitochondrial metabolism, AOX-expressing flies showed developmental problems (low rate of pupa eclosion) whose intensity appears to be related to the amount of carbohydrates (especially glucose) in the diet. Interestingly, we also found that strong expression of AOX in the fly testes causes low accumulation of mature spermatozoids, which in turn causes reproductive disadvantages. We are using a variety of molecular biology techniques (including RNA-Seq and high-resolution mass spectrometry) to investigate the changes in cell transcriptomics and mitochondrial proteomics and respiration, providing data about the physiological alterations caused by AOX and insights into the evolution of the mitochondrial alternative pathways in animals.
Figure 1. Phylogenetic relationships among animal taxa (according to [1]), indicating the lineages for which alternative enzyme genes have been found. Although vertebrates and arthropods are groups with the highest number of complete genomes sequenced (black circles), there is no evidence for the presence of the alternative oxidase (AOX) and the alternative NADH dehydrogenase (ANDH) in these animals. For taxa with no indications, such as Ctenophora, Nemertea, Platyhelminthes and Brachiopoda, there are not enough biochemical or genomic sequence data available to determine presence or absence of the alternative enzymes. Figure adapted from [2].
• O2k-Network Lab: FI Helsinki Jacobs HT, BR Jaboticabal Oliveira MT
Labels: MiParea: Respiration, Exercise physiology;nutrition;life style
Stress:Temperature Organism: Hexapods, Other invertebrates
Coupling state: OXPHOS
References
- Philippe H, Derelle R, Lopez P, Pick K, Borchiellini C, et al. (2009) Phylogenomics revives traditional views on deep animal relationships. Curr Biol 19: 706-712.
- Matus-Ortega MG, Salmeron-Santiago KG, Flores-Herrera O, Guerra-Sanchez G, Martinez F, et al. (2011) The alternative NADH dehydrogenase is present in mitochondria of some animal taxa. Comp Biochem Physiol Part D Genomics Proteomics 6: 256-263.
Affiliations and Support
Inst Biosc Med Tech, Univ Tampere, Finland. Mol Neurol Program, Univ Helsinki, Finland. Dept Tecnologia, Faculty Ciências Agrárias e Veterinárias, Univ Estadual Paulista “Júlio de Mesquita Filho”, Jaboticabal, SP, Brazil. - [email protected], Telephone: +55 1632097536.
This work has been supported by the Marie Curie International Incoming Fellowship, the Academy of Finland, and the Fundação de Amparo à Pesquisa do Estado de São Paulo.