Martell 2023 Nat Commun
| Martell E, Kuzmychova H, Kaul E, Senthil H, Chowdhury SR, Morrison LC, Fresnoza A, Zagozewski J, Venugopal C, Anderson CM, Singh SK, Banerji V, Werbowetski-Ogilvie TE, Sharif T (2023) Metabolism-based targeting of MYC via MPC-SOD2 axis-mediated oxidation promotes cellular differentiation in group 3 medulloblastoma. https://doi.org/10.1038/s41467-023-38049-9 |
» Nat Commun 14:2502. PMID: 37130865 Open Access
Martell Emma, Kuzmychova Helgi, Kaul Esha, Senthil Harshal, Chowdhury Subir Roy, Morrison Ludivine Coudiere, Fresnoza Agnes, Zagozewski Jamie, Venugopal Chitra, Anderson Chris M, Singh Sheila K, Banerji Versha, Werbowetski-Ogilvie Tamra E, Sharif Tanveer (2023) Nat Commun
Abstract: Group 3 medulloblastoma (G3 MB) carries the worst prognosis of all MB subgroups. MYC oncoprotein is elevated in G3 MB tumors; however, the mechanisms that support MYC abundance remain unclear. Using metabolic and mechanistic profiling, we pinpoint a role for mitochondrial metabolism in regulating MYC. Complex-I inhibition decreases MYC abundance in G3 MB, attenuates the expression of MYC-downstream targets, induces differentiation, and prolongs male animal survival. Mechanistically, complex-I inhibition increases inactivating acetylation of antioxidant enzyme SOD2 at K68 and K122, triggering the accumulation of mitochondrial reactive oxygen species that promotes MYC oxidation and degradation in a mitochondrial pyruvate carrier (MPC)-dependent manner. MPC inhibition blocks the acetylation of SOD2 and oxidation of MYC, restoring MYC abundance and self-renewal capacity in G3 MB cells following complex-I inhibition. Identification of this MPC-SOD2 signaling axis reveals a role for metabolism in regulating MYC protein abundance that has clinical implications for treating G3 MB.
• Bioblast editor: Plangger M • O2k-Network Lab: CA Winnipeg Banerji V
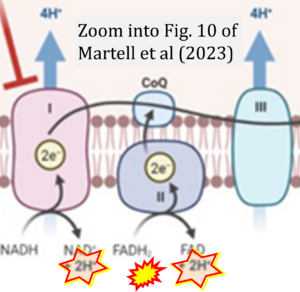
Correction: FADH2 and Complex II
- FADH2 is shown as the substrate feeding electrons into Complex II (CII). This is wrong and requires correction - for details see Gnaiger (2024).
- Gnaiger E (2024) Complex II ambiguities ― FADH2 in the electron transfer system. J Biol Chem 300:105470. https://doi.org/10.1016/j.jbc.2023.105470 - »Bioblast link«
Hydrogen ion ambiguities in the electron transfer system
Communicated by Gnaiger E (2023-10-08) last update 2023-11-10
- Electron (e-) transfer linked to hydrogen ion (hydron; H+) transfer is a fundamental concept in the field of bioenergetics, critical for understanding redox-coupled energy transformations.
- However, the current literature contains inconsistencies regarding H+ formation on the negative side of bioenergetic membranes, such as the matrix side of the mitochondrial inner membrane, when NADH is oxidized during oxidative phosphorylation (OXPHOS). Ambiguities arise when examining the oxidation of NADH by respiratory Complex I or succinate by Complex II.
- Oxidation of NADH or succinate involves a two-electron transfer of 2{H++e-} to FMN or FAD, respectively. Figures indicating a single electron e- transferred from NADH or succinate lack accuracy.
- The oxidized NAD+ is distinguished from NAD indicating nicotinamide adenine dinucleotide independent of oxidation state.
- NADH + H+ → NAD+ +2{H++e-} is the oxidation half-reaction in this H+-linked electron transfer represented as 2{H++e-} (Gnaiger 2023). Putative H+ formation shown as NADH → NAD+ + H+ conflicts with chemiosmotic coupling stoichiometries between H+ translocation across the coupling membrane and electron transfer to oxygen. Ensuring clarity in this complex field is imperative to tackle the apparent ambiguity crisis and prevent confusion, particularly in light of the increasing number of interdisciplinary publications on bioenergetics concerning diagnostic and clinical applications of OXPHOS analysis.
Labels: MiParea: Respiration, Genetic knockout;overexpression
Pathology: Cancer
Organism: Human Tissue;cell: Nervous system Preparation: Permeabilized cells, Intact cells Enzyme: Complex I, Complex II;succinate dehydrogenase
Coupling state: LEAK, ROUTINE, OXPHOS, ET Pathway: N, S, CIV, ROX HRR: Oxygraph-2k
2023-05