- high-resolution terminology - matching measurements at high-resolution
Electron-transfer-pathway state
Description
Electron-transfer-pathway states are obtained in mitochondrial preparations (isolated mitochondria, permeabilized cells, permeabilized tissues, tissue homogenate) by depletion of endogenous substrates and addition to the mitochondrial respiration medium of fuel substrates (CHNO) activating specific mitochondrial pathways, and possibly inhibitors of specific pathways. Mitochondrial electron-transfer-pathway states have to be defined complementary to mitochondrial coupling-control states. Coupling-control states require ET-pathway competent states, including oxygen supply. Categories of SUIT protocols are defined according to mitochondrial ET-pathway states. » MiPNet article
Abbreviation: ET-pathway state
Reference: BEC 2020.1, Gnaiger 2009 Int J Biochem Cell Biol, Gnaiger 2020 BEC MitoPathways, Categories of SUIT protocols
ET-pathway states
| Gnaiger E (2019) ET-pathway states. Mitochondr Physiol Network 2019-06-11 (last edit since 2016-11-08). |
Abstract: ET-pathway states are defined in mitochondrial preparations complementary to coupling-control states in mitochondrial physiology.
• O2k-Network Lab: AT Innsbruck Gnaiger E
Labels:
Preparation: Permeabilized cells, Permeabilized tissue, Homogenate, Isolated mitochondria, SMP
Coupling state: LEAK, OXPHOS, ET
Pathway: F, N, S, Gp, CIV, NS, Other combinations
HRR: Theory
ET-pathway levels
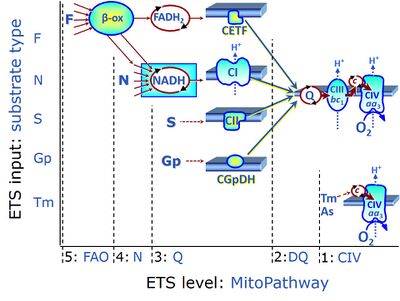
- ET-pathway levels are linked to ET-substrate types in mitochondrial SUIT protocols. Mitochondrial pathways are stimulated by fuel substrates (CHNO) feeding electrons into the Electron transfer pathway at different levels of integration and in the presence or absence of inhibitors acting on specific enzymes which control segments of these pathways. Distinction of five ET-pathway levels provides the rationale for defining categories of SUIT protocols.
- ET-pathway level 1 is stimulated by artificial electron donors (e.g. TMPD, Tm), reducing cytochrome c and feeding electrons to cytochrome c oxidase (CIV) or alternative oxidases, with oxygen as the terminal electron acceptor. When the CIV assay is carried out in the OXPHOS state, it may be considered as a short mitochondrial pathway of electron transfer or oxygen flux coupled to phosphorylation. In the noncoupled ET-state, however, the CIV assay measures the velocity of the single enzyme catalyzed step.
- ET-pathway level 2 is stimulated by duroquinol (DQ) feeding electrons into Complex III (CIII) with further electron transfer to CIV and oxygen.
- ET-pathway level 3 feeds electrons from NADH, FADH2, succinate, glycerophosphate into respiratory Complexes directly upstream of the Q-junction. NADH is the substrate of Complex I (CI). FADH2 is the substrate of electron transferring flavoprotein (CETF) localized on the inner side of the mt-inner membrane (mtIM). Succinate is the substrate of succinate dehydrogenase (SDH, CII) localized on the inner side of the mtIM. Glycerophosphate is the substrate of glycerophosphate dehydrogenase Complex (CGpDH) localized on the outer face of the mtIM. Choline and dihydro-orotate are the type 3 substrates of choline dehydrogenase and dihydro-orotate dehydrogenase, respectively.
- ET-pathway level 4 feeds electrons into dehydrogenases and enzyme systems upstream of pathway level 3. Electron transfer from NADH-linked substrates (N) converges at the N-junction. Representative N-junction substrates are pyruvate, glutamate and malate, and also citrate, oxoglutarate and others. The corresponding dehydrogenases (PDH, GDH, MDH and mtME; IDH, OgDH) generate NADH, the substrate of Complex I (CI). Fatty acids supporting converging electron transfer to the F-junction might be considered as type 4 substrates. However, the requirement of a combined operation of the F-junction and N-junction puts type F substrates to level 5 of pathway integration.
- ET-pathway level 5 feeds electrons into dehydrogenases and enzyme systems upstream of pathway level 3 with an obligatory combination of the F-junction and N-junction. F-junction substrates are fatty acids involved in β-oxidation, generating (enzyme-bound) FADH2, the substrate of electron transferring flavoprotein (CETF). Succinate does not belong to the type 4 substrates, since FADH2 is the product of CII, whereas FADH2 is the substrate of CETF. Fatty acid oxidation (FAO) not only depends on electron transfer through the F-junction (which is typically rate-limiting) but simultaneously generates NADH and thus depends on N-junction throughput. Hence FAO is inhibited by inhibition of Complex I (CI). In addition and independent of this source of NADH, the N-junction substrate malate is required as a co-substrate for FAO in mt-preparations, since accumulation of acetyl-CoA inhibits FAO in the absence of malate. Malate is oxidized in a reaction catalyzed by malate dehydrogenase to oxaloacetate (yielding NADH), which then stimulates the entry of acetyl-Co into the TCA cycle catalyzed by citrate synthase.
- Previously applied terms: ET-pathway substrate control state, Electron transfer-pathway state
ET-substrate types on different pathway levels
- ET-substrate type 5 on the pathway level of converging FADH2 and NADH-linked dehydrogenases, including beta-oxidation and segments of the TCA cycle:
- F: F-junction substrates, FADH2-linked, fatty acids (FAO)
- ET-substrate type 4 on the pathway level of converging NADH-linked dehydrogenases, including the TCA cycle:
- N: N-junction substrates, NADH-linked (and hence downstream 'CI-linked')
- ET-substrate type 3 on the pathway level of electron transfer Complexes converging at the Q-junction:
- Q: Q-junction substrates
- NADH, substrate of CI
- FADH2, substrate of CETF
- S: Succinate, substrate of CII
- Gp: Glycerophosphate, substrate of CGpDH
- Choline: substrate of choline dehydrogenase
- Dihydro-orotate: substrate of dihydro-orotate dehydrogenase
- ET-substrate type 1 on the single step level of cytochrome c oxidase (CIV), the terminal step in the aerobic electron-transfer pathway:
Terminology: from CI- and CII-linked to N and S
- CI and CII are abbreviations for enzymes, respiratory Complex I (CI) and Complex II (CII).
- In Gnaiger 2009, the 2007-2020 editions of Gnaiger 2020 BEC MitoPathways and many previous publications, CI-linked respiration has been used to indicate respiration supported by NADH-generating substrates, N (pyruvate, glutamate, malate, or other ET-pathway competent N-type substrate combinations). In this N pathway, electron transfer converges from dehydrogenases to the NADH-junction, and from NADH through CI to the Q-junction, with further electron transfer through CIII and CIV to oxygen. Similarly, CII-linked respiration indicates respiration supported by succinate, S, and electron transfer through CII to the Q-junction, with further electron transfer through CIII and CIV to oxygen.
- Convergent electron flow from a combination of NADH-generating substrates (N) and succinate (S) has been indicated as CI&II-linked respiration (Gnaiger 2020 BEC MitoPathways), synonymous with NS. The symbol '&' in CI&II helps to distinguish CI&II as the measured flux in the presence of both NADH-linked substrates and succinate, in contrast to CI+CII (or N+S) as the algebraic sum of fluxes measured separately in the N- versus S pathway control states.
- Simplification: NS denotes flux under the simultaneous control of the NADH- and succinate-linked pathways (NS-substrate cocktail), in contrast to N+S as the algebraic sum of the fluxes controlled by the two pathways separately. (Please, do not consider the symbol NS to indicate a multiplication, NS≠N·S.)
ET-pathway competent states
- In mitochondrial preparations coupling states (LEAK respiration, OXPHOS, ET-pathway) require ET-pathway competent states based on external substrate supply, including sufficient oxygen supply. ET-pathway competence of external substrates requires (i) transport of substrates across the mtIM or oxidation by dehydrogenases located on the outer face of the mtIM (e.g. glycerophosphate dehydrogenase Complex CGpDH), (ii) oxidation in the mt-matrix (TCA cycle dehydrogenases and other matrix dehydrogenases, e.g. mtGDH) or on the inner face of the mtIM (succinate dehydrogenase), (iii) oxidation of substrates without accumulation of inhibitory endproducts (e.g. oxaloacetate inhibiting succinate dehydrogenase; NADH and oxaloacetate inhibiting malate dehydrogenase), and (iv) electron transfer through the membrane-bound ET-pathway (mET-pathway). Endproducts must be either easily exported from the matrix across the mtIM (e.g. malate formed from succinate via fumarate), or metabolized in the TCA cycle (e.g. malate-derived oxaloacetate forming citrate in the presence of external pyruvate&malate).
Single electron-transfer-pathway states
- Single electron-transfer-pathway states are electron-transfer-pathway states for selective entry of electrons into the Q-junction through one particular respiratory Complex; for instance N-respiration through CI (PM; GM; PGM with or without malonic acid: Gnaiger 2020 BEC MitoPathways), S-electron-transfer-pathway state with e-input into Q through CII, CIV (Tm: MiPNet06.06_Chemical O2 background).
- Further details, see Categories of SUIT protocols.
Multiple electron-transfer-pathway state
- Multiple electron-transfer-pathway states are electron-transfer-pathway states obtained in living cells respiring on endogenous substrates or in media with physiological exogenous substrates, or designed for reconstitution of TCA cycle function in isolated mitochondria, permeabilized cells or permeabilized tissues. In all cases, electron flow converges at the Q-junction with multiple entry sites of electron transfer in the NS pathway through CI&II, FNS pathway through FAO&CI&II, NSGp pathway through CI&II&GpDH.
- Further details » Categories of SUIT protocols
Pathway versus kinetic substrate control
Control by substrate type: electron-transfer-pathway state
- A: Living cells
- Endogenous pathway control: In living cells, ce, endogenous organic carbon substrates are mobilized in the cytosol as intermediary metabolites transported across the mtIM and thus exerting control over mitochondrial respiration. If no organic carbon substrates are supplied in the incubation medium, then substrate control is entirely endogenous. Long-term incubation under such conditions leads to progressive depletion of endogenous substrates.
- Exogenous pathway control: Cells are grown in complex culture media with a variety of organic carbon substrates, and different exogenous electron-transfer-pathway states are achieved by variation of these substrates. Long-term incubation in closed systems without exchange of culture medium leads to progressive depletion of exogenous substrates. Incubation of cells in simple media allows for sequential titration of specific carbon substrates (e.g. glucose or fructose; lactate or glutamate; fatty acids) for the study of exogenous pathway control of respiration.
- A: Living cells
- B: Mitochondrial preparations
- Specific substrate-inhibitor combinations are selected to establish pathway states for (i) stimulating defined segments of the electron-transfer pathway, or (ii) reconstitution of TCA cycle function.
- B: Mitochondrial preparations
- Electron-transfer-pathway state with electron gating: Specific substrate-inhibitor combinations are applied for selectively stimulating electron entry from N-junction substrates through CI, S-Electron-transfer-pathway state through CII, or other substrates feeding additional branches converging at the Q-junction, particularly F-type (fatty acid oxidation and Gp (glycerophosphate). The most commonly applied pathway states select for N-electron input through Complex I (pyruvate&malate, PM; glutamate&malate, GM), S-electron-transfer-pathway state ([[Complex II] pathway to Q]: succinate and rotenone, SRot), or Complex IV electron input (CIV: ascorbate&TMPD(Ama)).
- Physiological electron-transfer-pathway states: Reconstitution of TCA cycle function requires NS-substrate combinations, such as PMS, GMS, GS, PGMS, or PGS, applied simultaneously without inhibitor of any respiratory Complexes.
Control by substrate concentration: kinetic control states
- Kinetic substrate or adenylate control: Kinetic studies with variation of a specific substrate (reduced substrate supplying electrons to the ET pathway; ADP, Pi; O2; cytochrome c) are analyzed by kinetic functions (e.g. hyperbolic), yielding kinetic parameters, such as Jmax, Km', c50 [µM], or p50 [kPa].
- Kinetic inhibitor control: Kinetic studies with variation of a specific inhibitor yield apparent kinetic constants, such as the KI'.
Related MitoPedia pages
- Electron-transfer pathway, ET pathway
- ET capacity
- Coupling-control state E
References
- Bioblast links: Uncoupling - >>>>>>> - Click on [Expand] or [Collapse] - >>>>>>>
- Specific
- » Artefacts by single dose uncoupling
- » ATP synthase
- » CCCP
- » Coupling-control protocol
- » DNP
- » Dyscoupled respiration
- » FCCP
- » Is respiration uncoupled - noncoupled - dyscoupled?
- » Noncoupled respiration: Discussion
- » Uncoupler
- » Uncoupled respiration - see » Noncoupled respiration
- » Uncoupling proteins
- » Uncoupling protein 1
- » Uncoupler titrations - Optimum uncoupler concentration
- Specific
- Respiratory states and control ratios
- » Biochemical coupling efficiency
- » Coupling-control state
- » Electron-transfer-pathway state
- » Electron-transfer pathway
 ET capacity
ET capacity- » E-L coupling efficiency
- » Flux control efficiency
- » Flux control ratio
- » LEAK-control ratio
- » LEAK respiration
- » Noncoupled respiration
- » OXPHOS
- » OXPHOS capacity; » State 3
- » OXPHOS-control ratio, P/E ratio
- » Respiratory acceptor control ratio
- » ROUTINE-control ratio
- » ROUTINE respiration
- » ROUTINE state
- » State 3u
- » State 4
- » Uncoupling-control ratio UCR
- Respiratory states and control ratios
- Gnaiger E et al ― MitoEAGLE Task Group (2020) Mitochondrial physiology. Bioenerg Commun 2020.1. https://doi.org/10.26124/bec:2020-0001.v1
- Gnaiger E (2020) Mitochondrial pathways and respiratory control. An introduction to OXPHOS analysis. 5th ed. Bioenerg Commun 2020.2. https://doi.org/10.26124/bec:2020-0002
- General (alphabetical order)
- Other keyword lists
MitoPedia concepts:
MiP concept,
Respiratory state,
SUIT concept,
SUIT state,
Recommended
MitoPedia methods:
Respirometry
MitoPedia topics:
Substrate and metabolite,
EAGLE